DeepREx-WS
A Deep-learning based method for the prediction of Residue solvent Exposure starting from protein sequence
New version available here based on Protein Language Models for faster and more accurate predictions!
Using this webserver
From the Home page of this website, the user can submit one input sequence in fasta format to the predictor by copying the text in the panel and pressing the button "Submit".
In order to be able to submit a job, you must specify an ID line starting with a '>' character, followed by the protein sequence, that can have between 50 and 5000 valid residues (ARNDCQEGHILKMFPSTWYVUXZB). If any problem is found in your input, the box will turn red and it will notify you of what you should change, when the box becomes green it means everything is okay and you can press the button "Submit".
If you want to test the predictor, you can click on the button "Load example FASTA" and submit that sequence, the results will be available in a few minutes.
If you need to run predictions on a large dataset of proteins, we suggest that you download the standalone version of this program (instructions below).

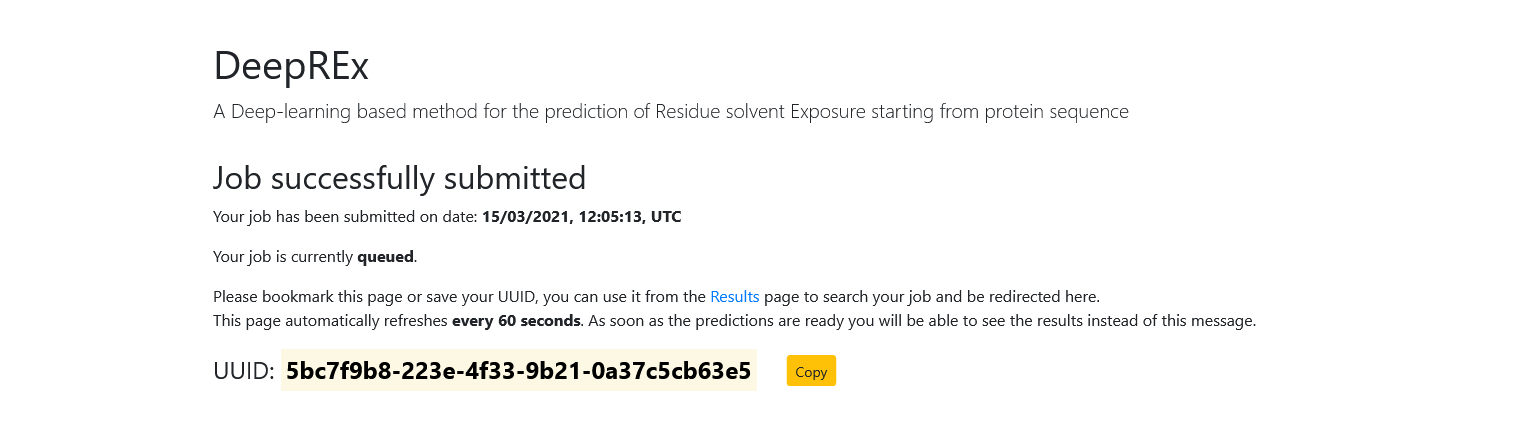
Once you submit a query, you will be redirected to a new page. This means that your job has been accepted and you need to wait for the results. This page automatically refreshes every 60 seconds and it will display the status of your job (either queued or running).
In the meantime, you should both bookmark the page (the results will be available at the same URL once they are ready) and save the Universally Unique IDentifier of your job using the "Copy" button.
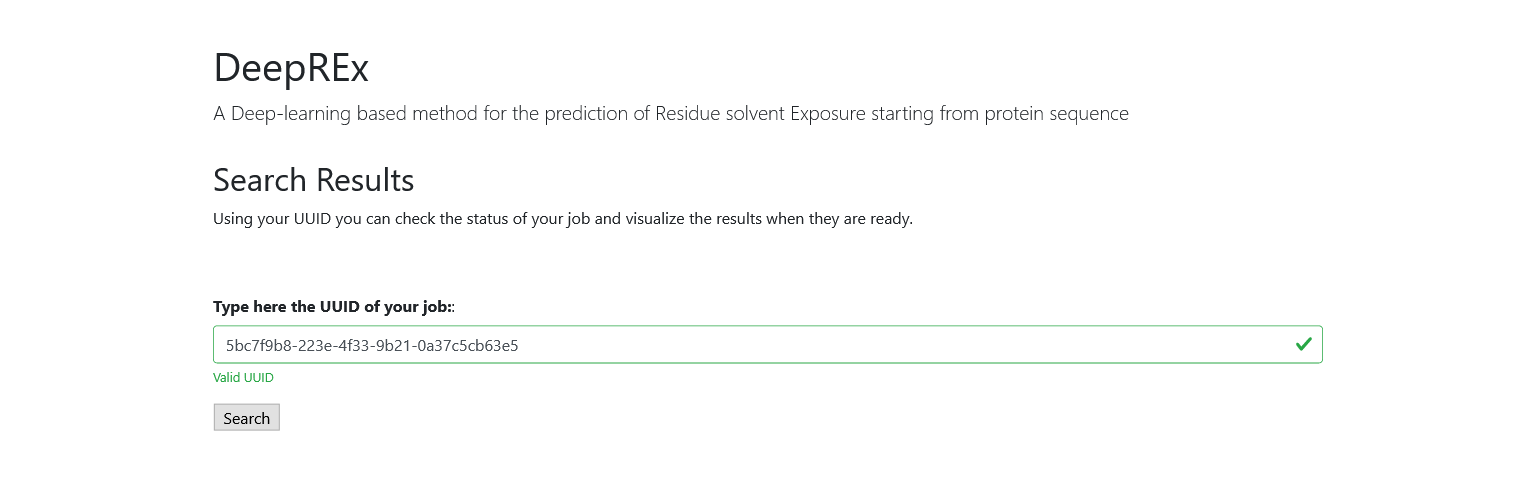
You can then use the UUID from the Results page to search for your job at any time. Like in the Home page, you can only start the search with a valid id and you will be notified if it is not present in our database.
Reading the results
Once the results are ready, they will be made available to your result page. There are three main sections in this page:
- Job information
- Feature Viewer
- Data Tables
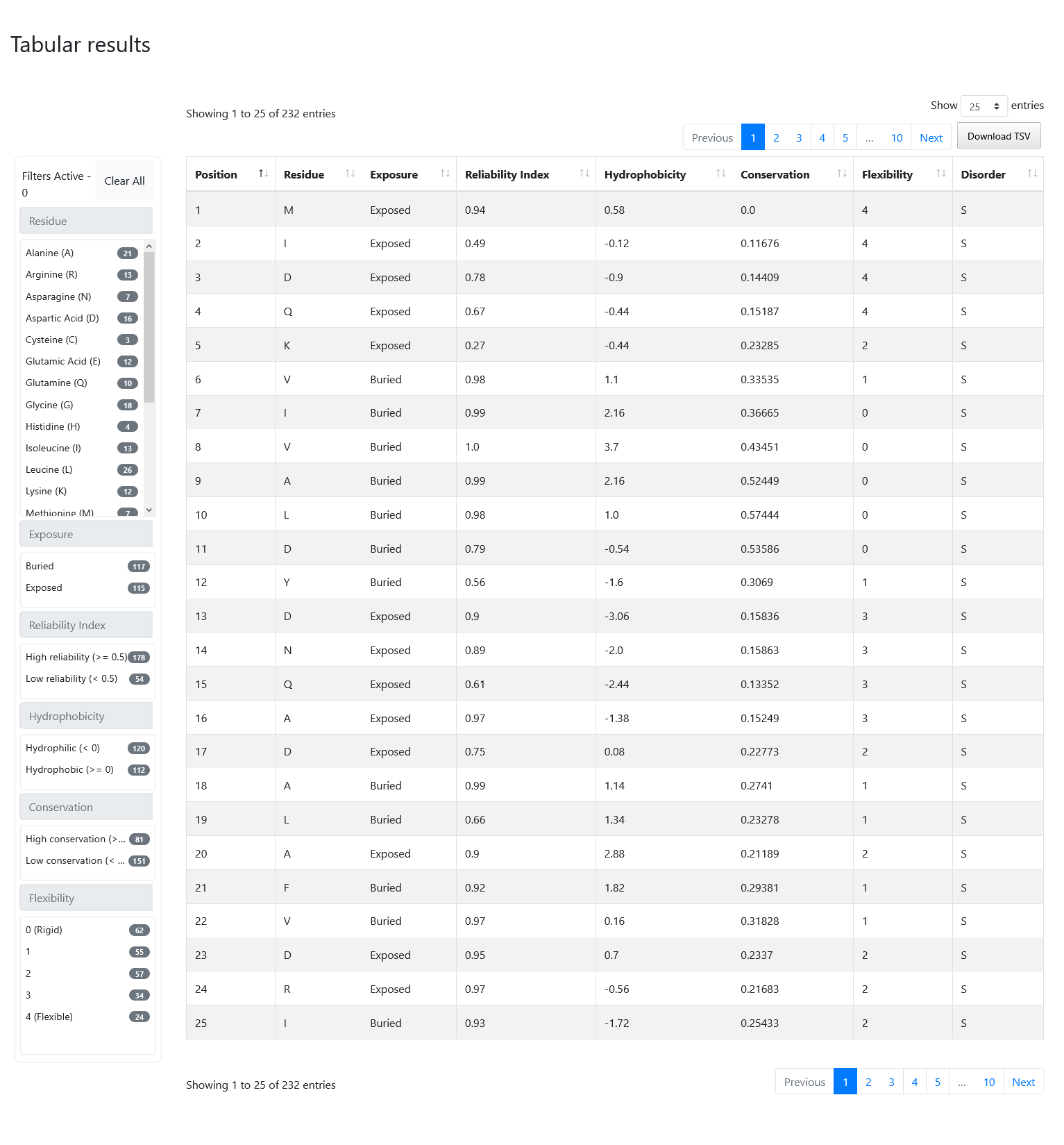
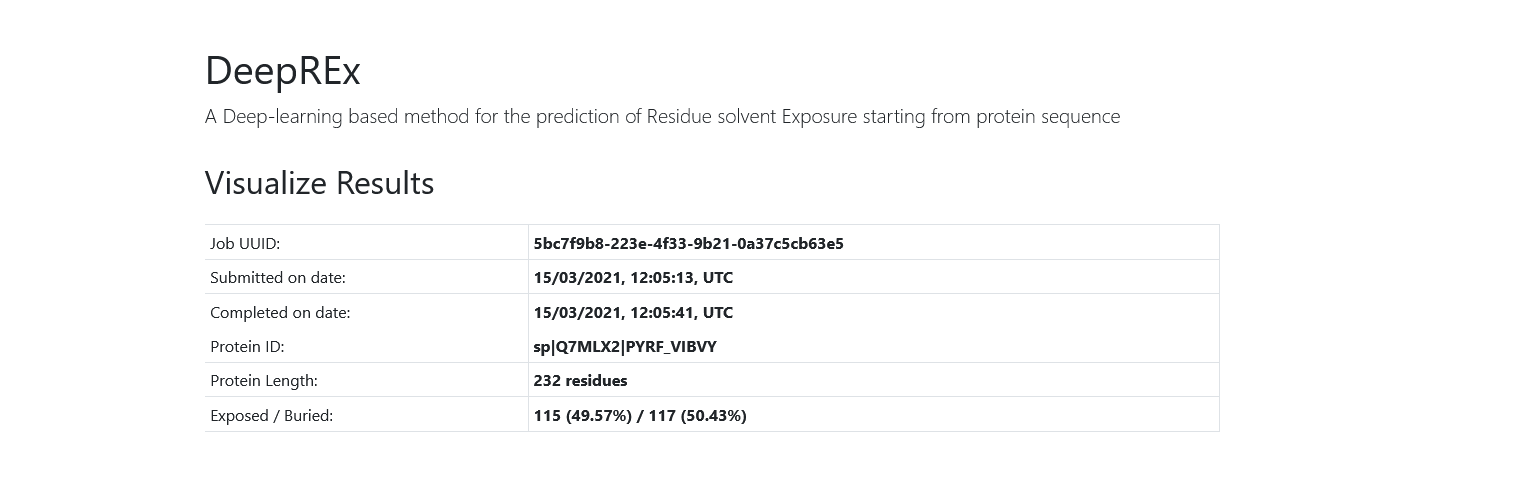
On the top of the page you will see some general information about your job, including your UUID, the date of submission and completion, the protein ID, the protein length and the counts and percentages of exposed vs buried predictions.
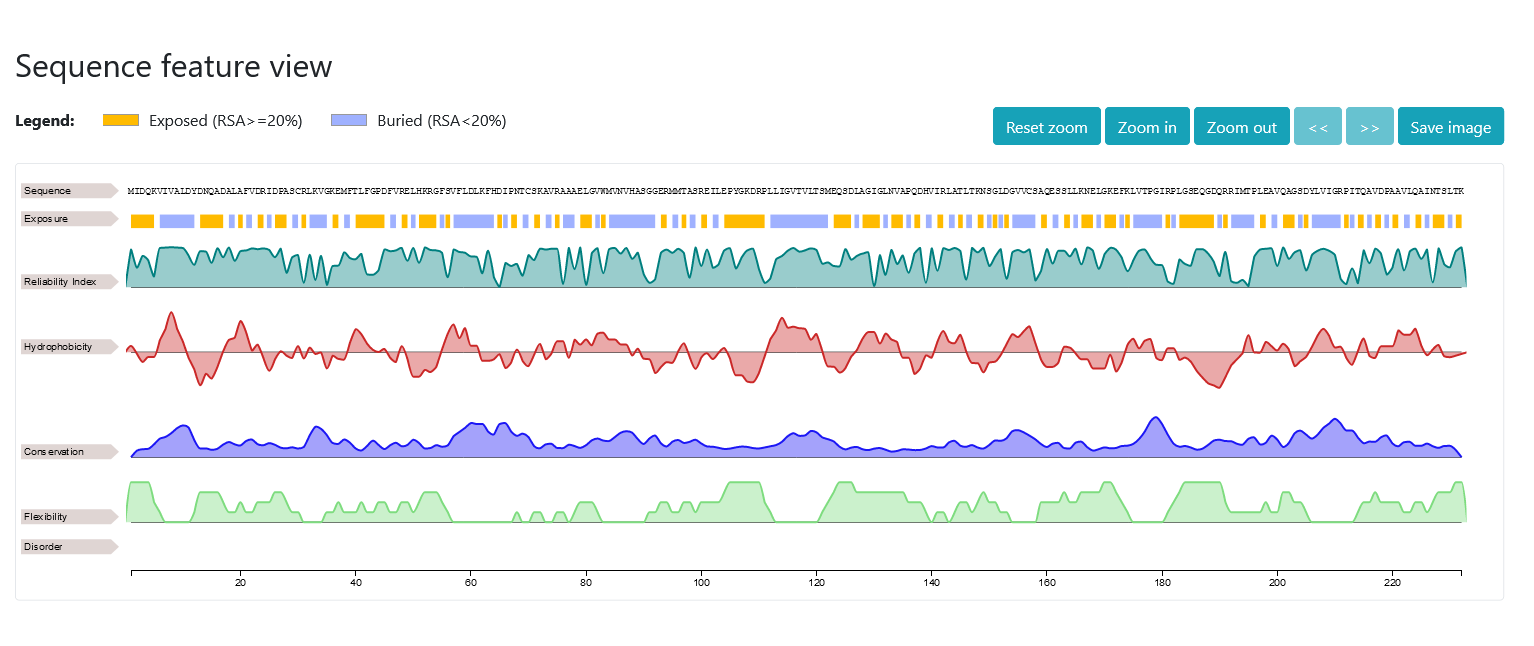
Right after, you can visualize the results with neXtProt feature viewer. The first line simply displays the residues of the sequence; the second and third lines show the output of DeepREx-WS, that is the exposure predictions and the corresponding reliability indices (real values between 0 and 1). In the remaining four lines, additional residue-level features correlated with the solvent accessibility are visualized. Those are in order: i) Kyte & Doolittle hydropathy scores averaged over a window of five residues (real values between -4.5 and +4.5), ii) conservation indices of the residues in the multiple sequence alignment (real values between 0 and 1), iii) predicted protein flexibility as obtained with Medusa (integer classes between 0 and 4) and iv) predicted intrinsically disordered regions as obtained with MobyDB-lite3.0 (colored boxes visualized on disordered regions).
If you hover the mouse over the graphs, the position of the residue will be shown. If you select a rectangular area, you can zoom on a part of the sequence. You can always right click on the graph to reset the zoom back to normal. Alternatively, you can make use of the buttons on the top-right of the viewer to zoom-in, zoom-out, move along the sequence or take a screenshot of the selected area.
At the bottom of the screen, you can visualize the exact same information described before inside a formatted table. On the left of the table you can see a number of filters that you can select and combine together. On the top-right of the table, there is a button called "Download TSV" that you can press to obtain the results displayed in the table on a Tab-Separated-Value format file, with an additional column on the left containing the protein ID in case you want to combine multiple files together. Note that if you applied any filters, you will only download the portion of the results that is currently selected.